Abstract
INTRODUCTION:
Atypical hemolytic uremic syndrome (aHUS) is a rare, genetic, progressive, life-threatening form of thrombotic microangiopathy (TMA). A genetic mutation has been found as an explanation for constitutive complement activation in 50-60% of cases. In this abstract, we present whole exome analysis data results and their clinical relevance in 12 patients with aHUS.
MATERIALS AND METHOD:
We have enrolled 24 patients in this study who were diagnosed as aHUS and followed in our clinic since 2012. For exome sequencing, libraries were prepared using SeqCap EZ Human Exome Library v3.0 kit. Real Time Analysis software version with standard parameters was used for real-time image analysis and base calling. Raw sequencing data were aligned to the human genome hg19 reference (Homo sapiens (hg19) sequence) using CLC Genomic Workbench V10 (Qiagen) with standard parameters in paired-end mode. All variants were filtered according to the following criteria: not in dbSNP144 and also 1000 genome MAF <0.01, homozygous in the affected patients and heterozygous in the carrier parents, Genotype Quality (GQ) >30 and coverage (DP) >20 and, in homozygous region.
FINDINGS AND RESULTS:
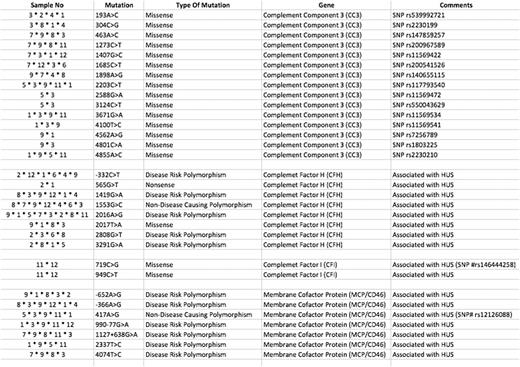
We performed whole exome sequencing in all 24 patients but during raw data analysis 12 patients were found to be eligible for further analysis according to QC value. We didn't find any mutation in one of the patients (case 10) who had presented with diarrhea and had very good response to plasmapheresis only. We detected 32 different complement mutations in 4 different genes including CC3, CFI, CFH and membrane MCP in all other 11 patients. These mutations are summarized in the table.
Four of the patients have responded to plasmapheresis and eculizumab was not needed. Of the remaining 20 patients eculizumab was aimed to be used. However, two of them died due to acute respiratory distress and thrombosis prior to eculizumab therapy. Case 9 was one of them and we found 21 different mutation including nine in CC3, five in CFH and 7 in MCF. One of our patients died in her 3rd month of eculizumab therapy (Case 2) and she had also seven mutations mostly in CFH gene. We used eculizumab in 18 of the patients and got complete response for thrombotic microangiopathy (TMA) in all. We had very good response to eculizumab in all these patients; even hemodialysis need disappeared in nine of them during eculizumab therapy. We experienced recurrence of renal failure in one patient (Case11) under eculizumab treatment in two years. She had also two different mutations in CFI gene four of the seven mutations in MCP gene and four of the 15 mutations in CC3 gene. This patient was lost due to multiple organ failure.
We decided to stop eculizumab at the ninth month of treatment in two of the patients (Case 1 and Case 3) who had complete hematologic and renal recovery. Case 1 relapsed 16 months later and successfully retreated with eculizumab. She had 15 different mutations as eight mutations in CC3, six mutations in CFH, and 1mutation in MCP gene. Case 3 is still under control for three years after stopping eculizumab treatment. But as an interesting finding, this patient was 80 years old and had the heaviest mutation load with 22 different mutations (6 of the 7 mutations in MCP gene, 11 of the 15 mutation in CC3 gene, 4 of the 8 mutation in CFH gene).
AHUS has been diagnosed during pregnancy in two of the patients and they had a successful delivery with healthy babies without any complication including ESRD. We also found 14 different mutations mostly in CFH gene in one of them (Case 8).
DISCUSSION AND CONCLUSION:
We found at least four different mutations in all patients except one case. Although MCP mutations accounts for only 15% of the aHUS cases we also found at least one MCP mutation in 91% of our cases. We found the rarest mutations in CFI gene. Although there are no clinical predictors of outcome, knowledge of the underlying genetic defect is helpful in predicting prognosis. But the patient who had the most number of mutations also had the best prognosis in our cohort. Therefore it is hard to use mutation data for management and prognosis of the syndrome. But the presence of a mutation is clearly important for the ESRD before transplantation decision and also would encourage patients to continue treatment.
Yenerel: Alexion: Honoraria. Caliskan: Alexion: Honoraria. Yildiz: Alexion: Honoraria. Turkmen: Alexion: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal